Tautomerization of 2-naphthol-AlCl3 complex
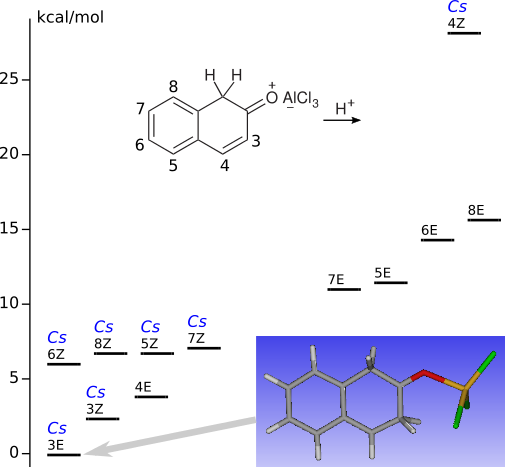
C-Protonation of 2-naphthol-AlCl3 complexes CE and CZ
Reaction of 2-naphthol-AlCl3 complex with benzene
Binol-AlCl3 complexes
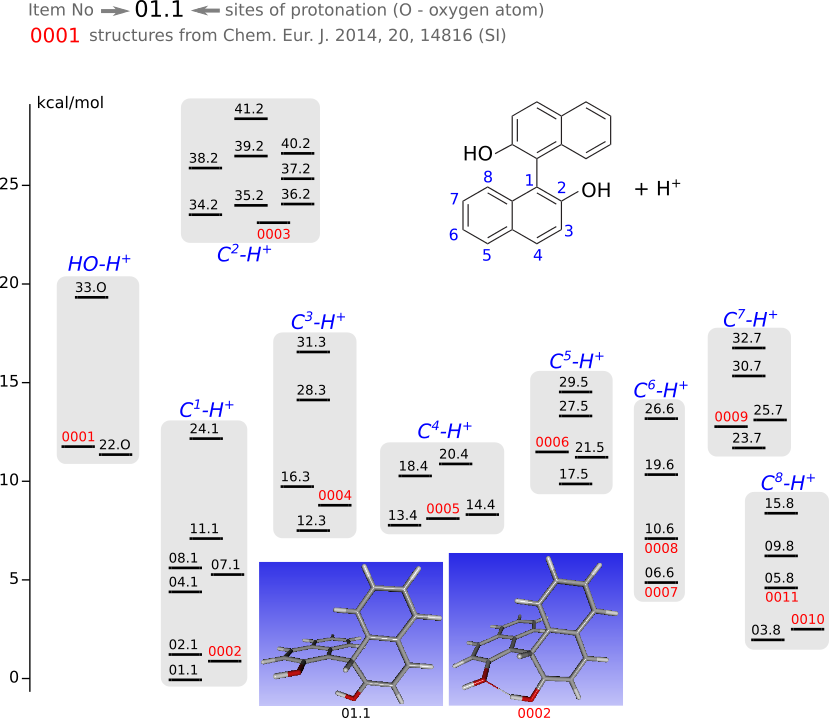
Protonated BINOL
Diprotonated BINOL
Protonated CO-tautomer of Binol-AlCl3 complex
Transition states of racemization of BINOL
Tautomerization of 2-naphthol-AlCl3 complex
The geometries of prereaction comlexes and transition states were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
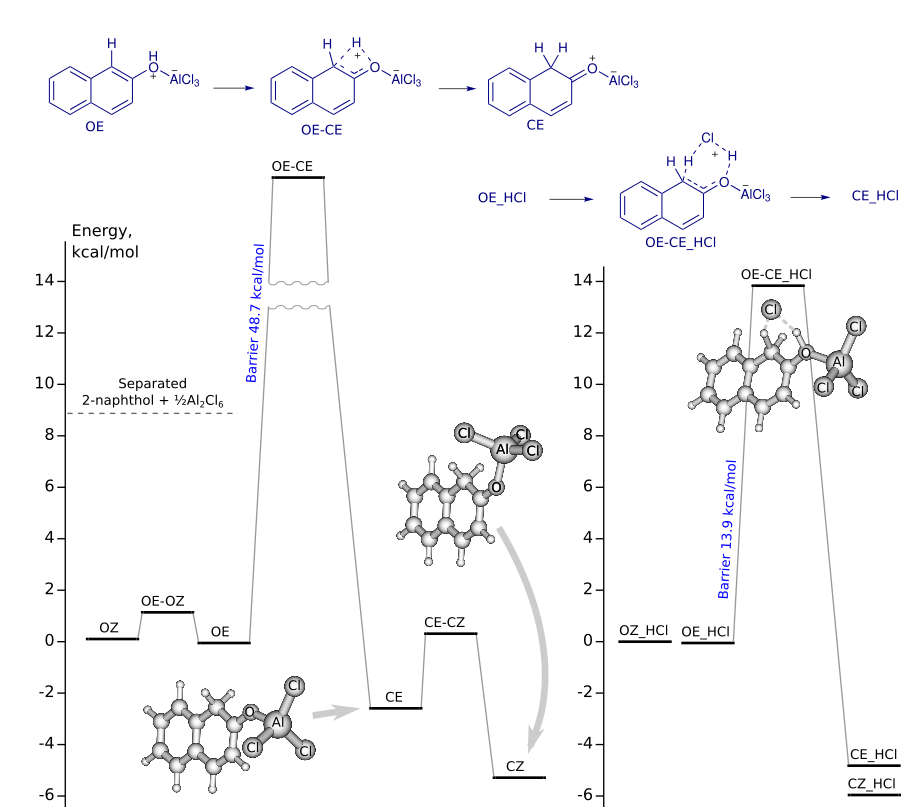
IRC (concatenated xyz) from OE-CE and OE-CE_HCl
C-Protonation of 2-naphthol-AlCl3 complexes CE and CZ
Reaction of 2-naphthol-AlCl3 complex with benzene
Conformational analysis of the reaction products (Ph-E and Ph-Z) was performed with the computer program package ChemAxon Marvin (conformers plugin).The geometries of all the conformers. prereaction comlexes and transition states were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
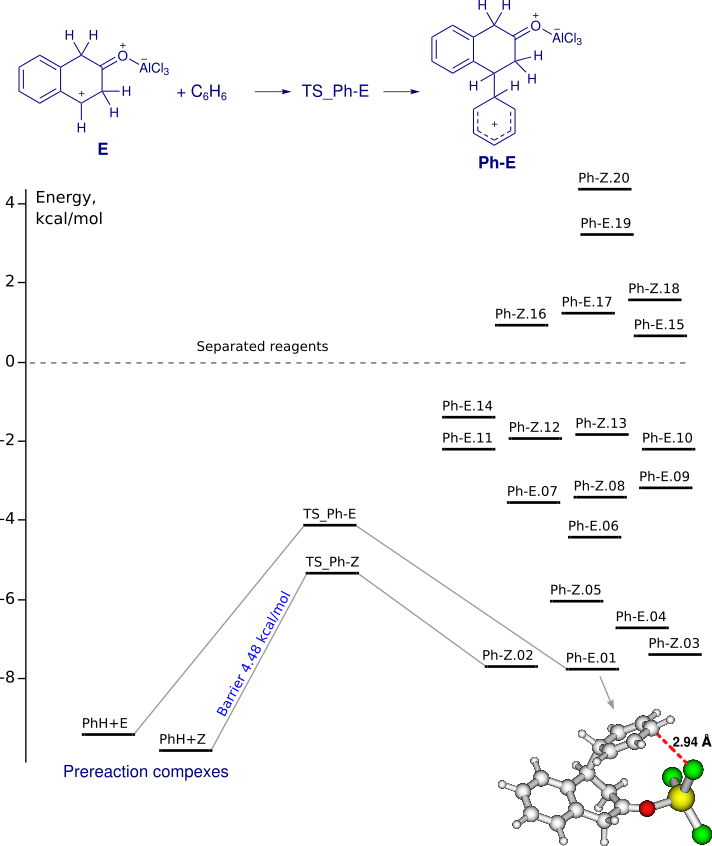
IRC (concatenated xyz) from TS_Ph-E and TS_Ph-Z
Binol-AlCl3 complexes
Binol is 1,1′-bi-2-naphtholConformational analysis of the complexes was performed with the computer program package ChemAxon Marvin (conformers plugin).
The geometries of all the conformers were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|

See also protonation of CO-tautomer
Protonated BINOL
The geometries of all the protonated forms were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
Diprotonated BINOL
The geometries of all the protonated forms were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
Yellow highlighted are structures from Chem. Eur. J. 2014, 20, 14816 (SI)
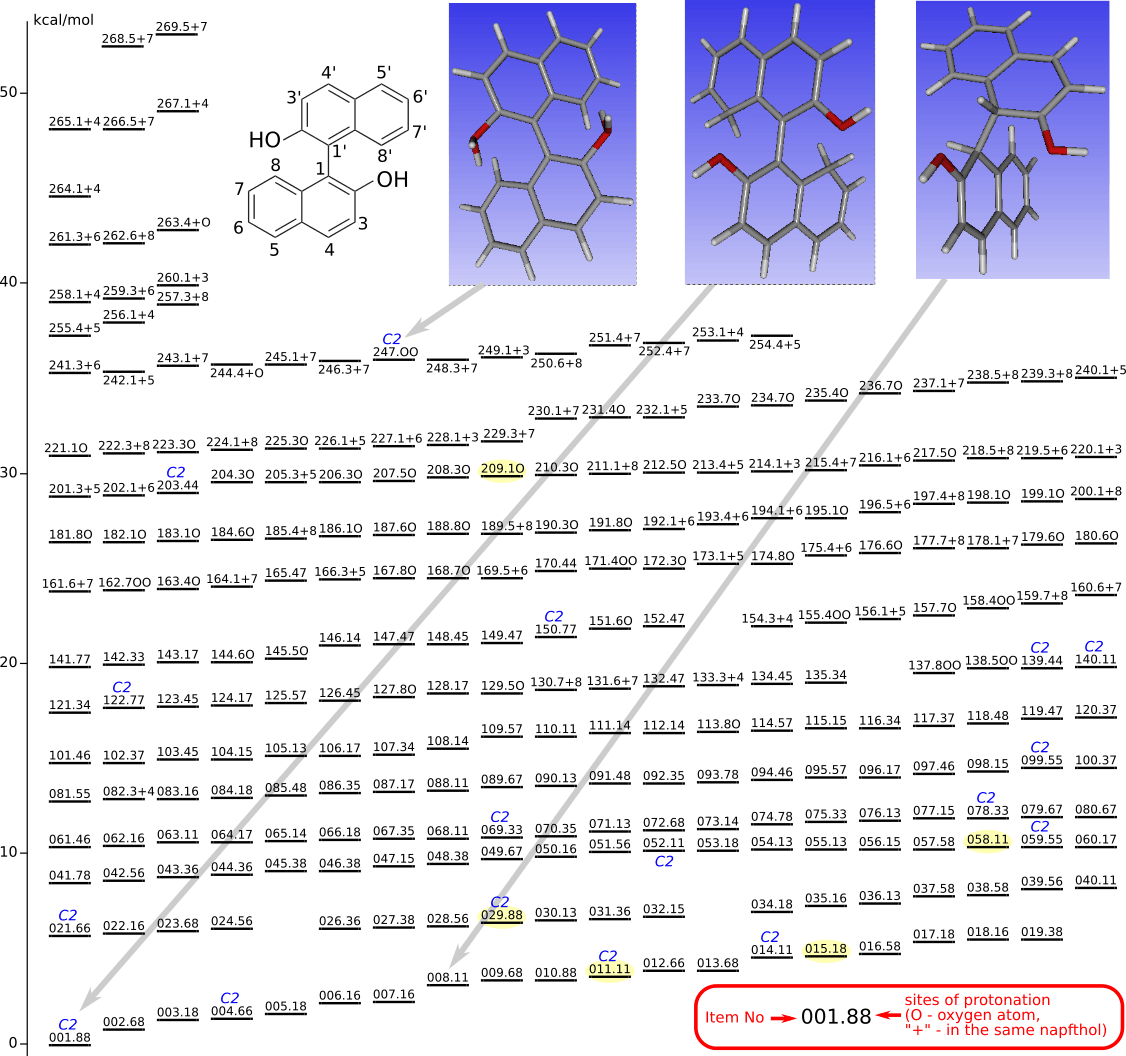
Relaxation of 247.OO into 010.88

IRC (concatenated xyz)
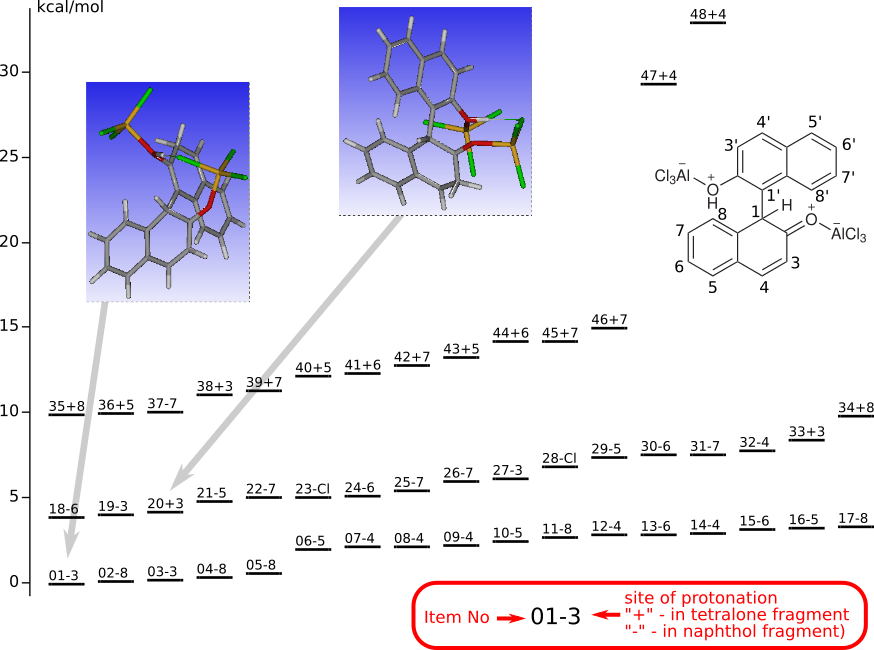
Protonated CO-tautomer of Binol-AlCl3 complex
Tautomers of Binol-AlCl3 complexThe geometries of all the protonated forms were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
Transition states of racemization of BINOL
The geometries of all the protonated forms and transition states were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
Energy level titles:
first digit — charge of the structure
second letter "l" — structure from Chem. Eur. J. 2014, 20, 14816 (SI) optimized by DFT/PBE/L1
suffix "TS" — transition state of racemization
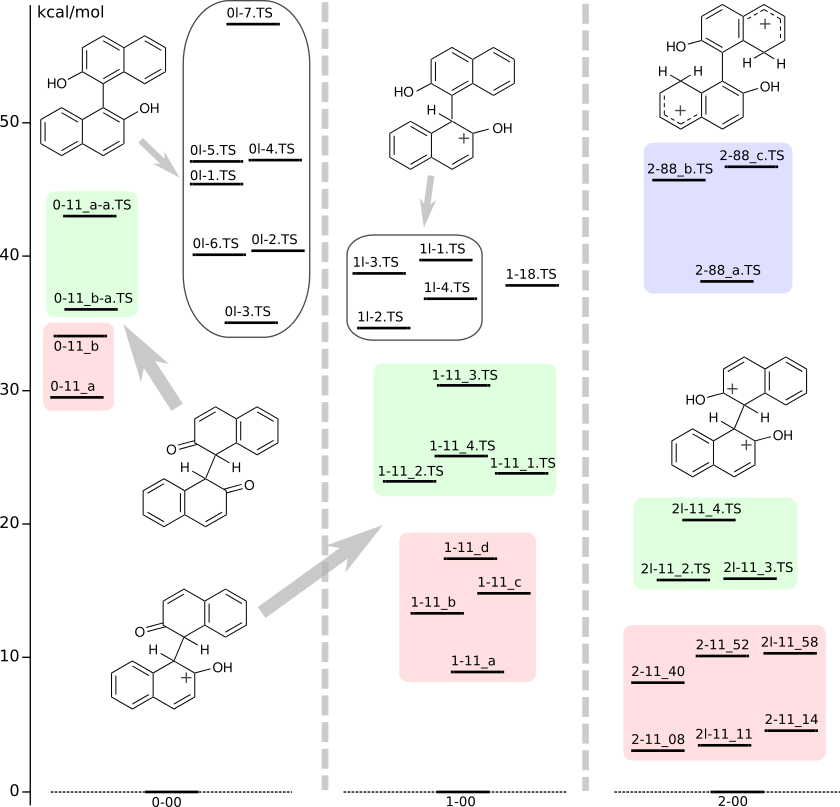
Zero energy levels: 0-00 — ZZ-BINOL, 1-00 — 01.1, 2-00 — 001.88
IRC (concatenated xyz): 0-11_b-a, 1-11_d-a (not racemization), 2l-1 (not racemization), 2-88_a
Rotations in 1,1'-diprotonated BINOLs
The geometries of all the protonated forms and transition states were optimized by DFT (Priroda program, PBE functional, L1 basis (Λ01, cc-pVDZ analog)).
| 
|
Correspondence with previous diagram:
11R and 11S are 008.11, 9R is 011.11, 9S is 014.11
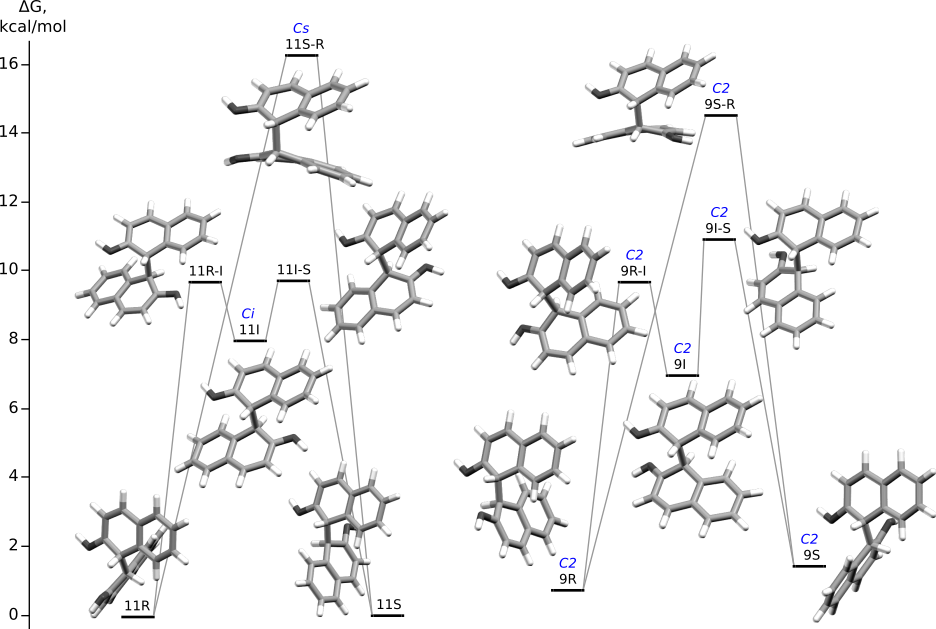
IRC (concatenated xyz): 11R-11S, 9R-9S