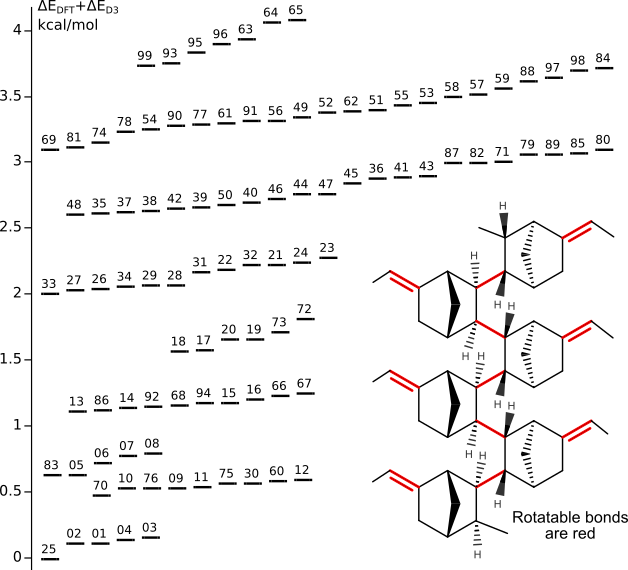
Conformers of ENB oligomer (as model of syndiotactic PENB)
Tinker scan (MMFF94 force field, energy window 20 kcal/mol) has generated 892 conformers.99 most stable conformers have optimized by DFT/PBE/L1 (PRIRODA program).
Energy and chemical shifts have calculated at DFT/PBE/L22 level.
Grimme's D3 dispersion corrections are for optimized geometries.
| 
|
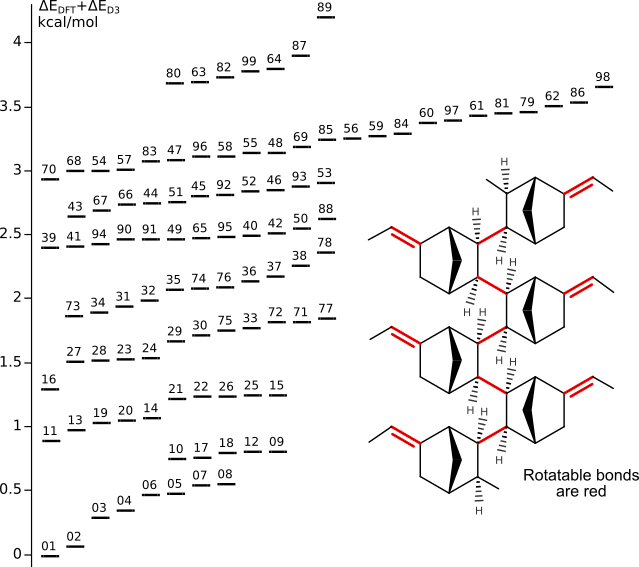
Conformers of ENB oligomer (as model of isotactic PENB)
Tinker scan (MMFF94 force field, energy window 20 kcal/mol) has generated 11473 conformers.99 most stable conformers have optimized by DFT/PBE/L1 (PRIRODA program).
Energy and chemical shifts have calculated at DFT/PBE/L22 level.
Grimme's D3 dispersion corrections are for optimized geometries.
Isotactic conformers are 26 kcal/mol less stable than syndiotactic ones.
| 
|
02 Sept 2019