|
|
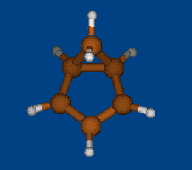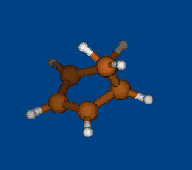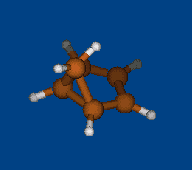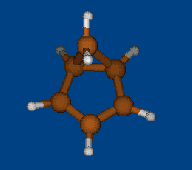
Conformational analysis of [1.1.1] cryptand
Methods: MM (Marvin+Vconf+Tinker) — RM1 (MOPAC2009) — DFT (PRIRODA).
Energy (without ZPE) by DFT/PBE/L1 (cc-pVDZ) geometry (PRIRODA program).
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|

Conformational analysis of [Li+⊂ 1.1.1] cryptate
Methods: MM (Marvin) — PM6 (MOPAC2009) — DFT (PRIRODA).
Energy (without ZPE) by DFT/PBE/L1 (cc-pVDZ) geometry (PRIRODA program).
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|

|