Новосибирский институт органической химии им. Н.Н. Ворожцова СО РАН
Лаборатория изучения механизмов органических реакций
 |
 |
 |
 |
 |
 |
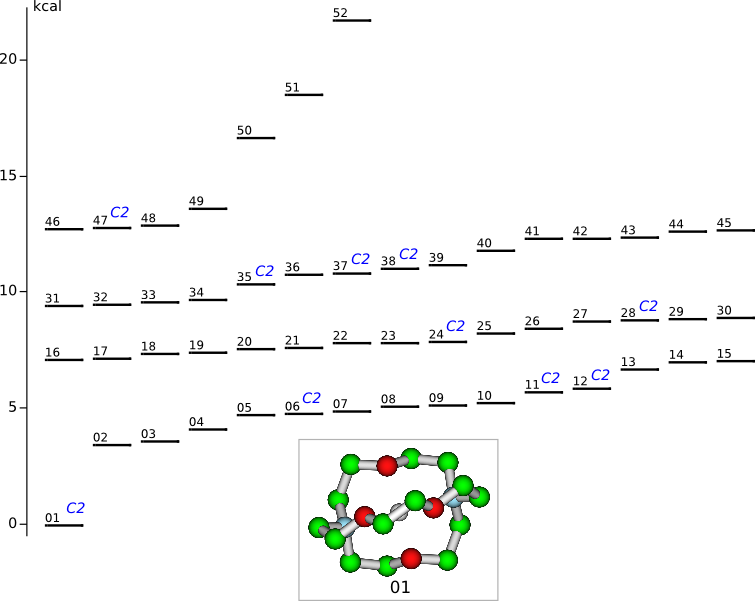
Conformational analysis of 2.1.1-cryptand
(4,7,13,18-tetraoxa-1,10-diazabicyclo[8.5.5]icosane)
Methods: MM (Marvin+Vconf+Tinker) — RM1 (MOPAC2009) — DFT (PRIRODA).Energy (without ZPE) and geometry by DFT/PBE/L1 (cc-pVDZ) (PRIRODA program).
Click to energy level to view 3D structure (run Jmol java-applet). Click to level inscription to download or open xyz coordinates file.

Conformational analysis of 2.1.1-cryptand Li+ complex
Methods: MM (Marvin) — PM6 (MOPAC2009) — DFT (PRIRODA).Energy (without ZPE) and geometry by DFT/PBE/L1 (cc-pVDZ) (PRIRODA program).
Click to energy level to view 3D structure (run Jmol java-applet). Click to level inscription to download or open xyz coordinates file.