|
|
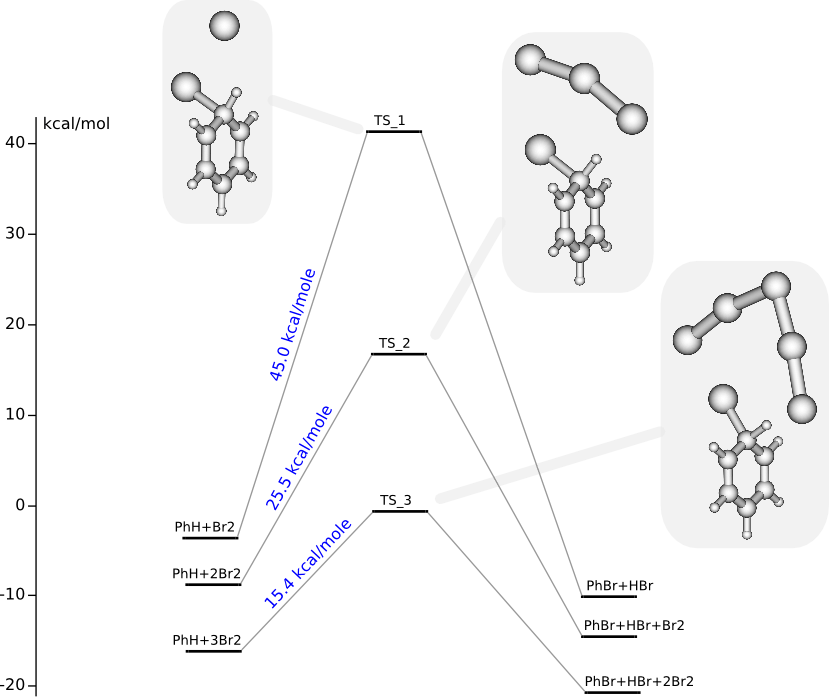
C6H6 + nBr2 ➙ C6H5Br + HBr + (n-1)Br2 (n = 1, 2, 3)
See Henry Rzepa blog.
Geometry optimization by DFT/PBE/L1 (PRIRODA program, basis like cc-pVDZ)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
Summary energy of noninteracting reagents is taken to be zero energy.
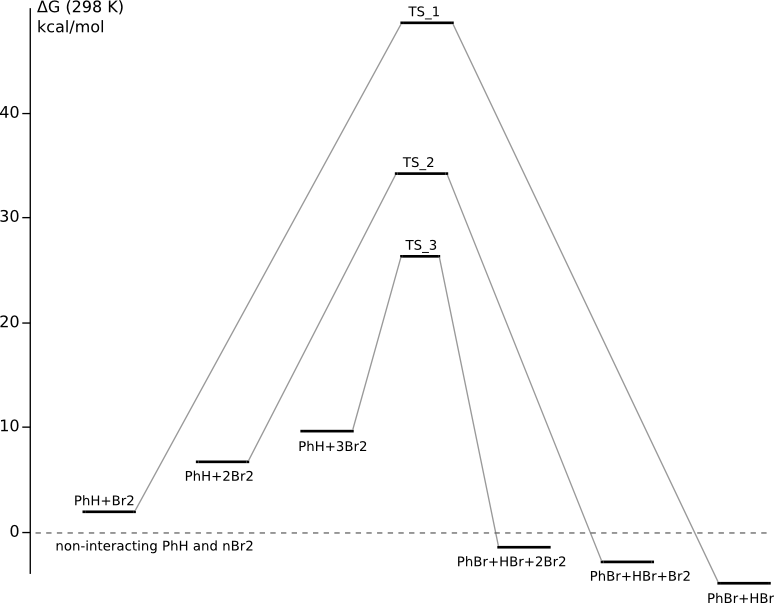
With thermochemistry corrections
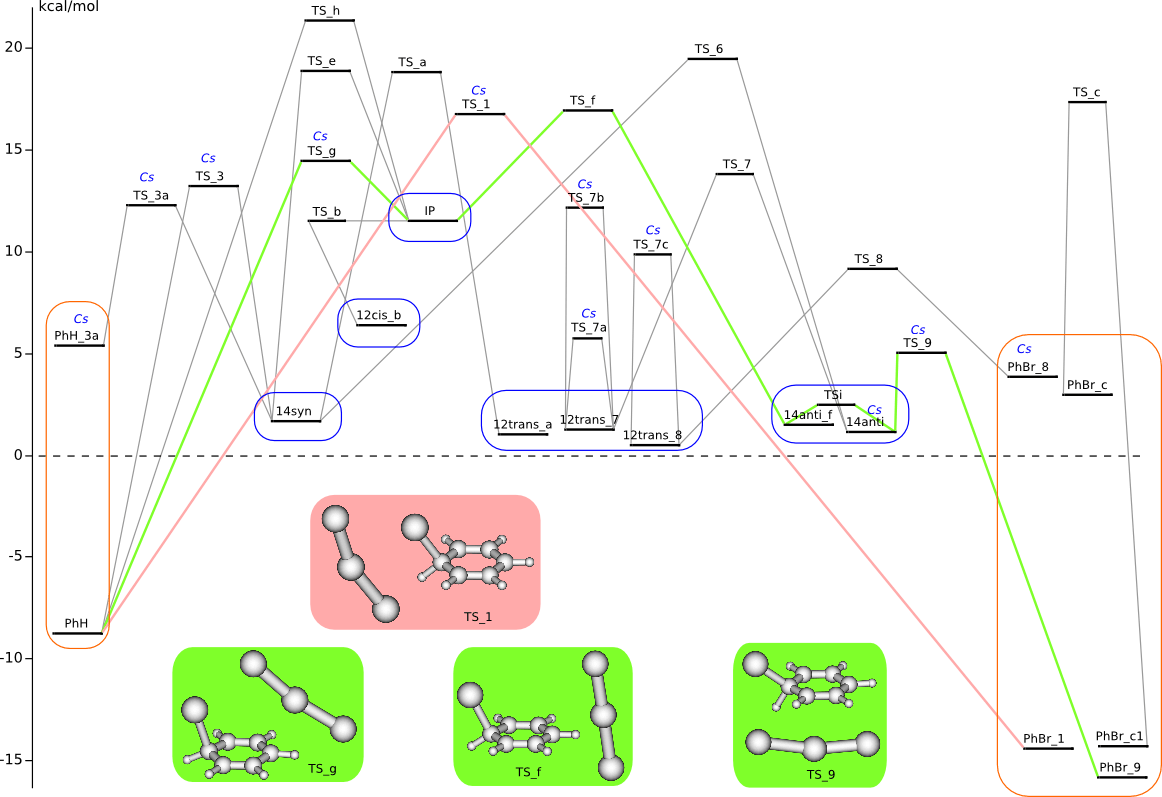
C6H6 + Br2 PES
cf.
J. Kong, B. Galabov, G. Koleva, J.-J. Zou, H. F. Schaefer III, and P.v.R. Schleyer, Angew. Chem. Int. Ed. 2011, 50, 6809-6813
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
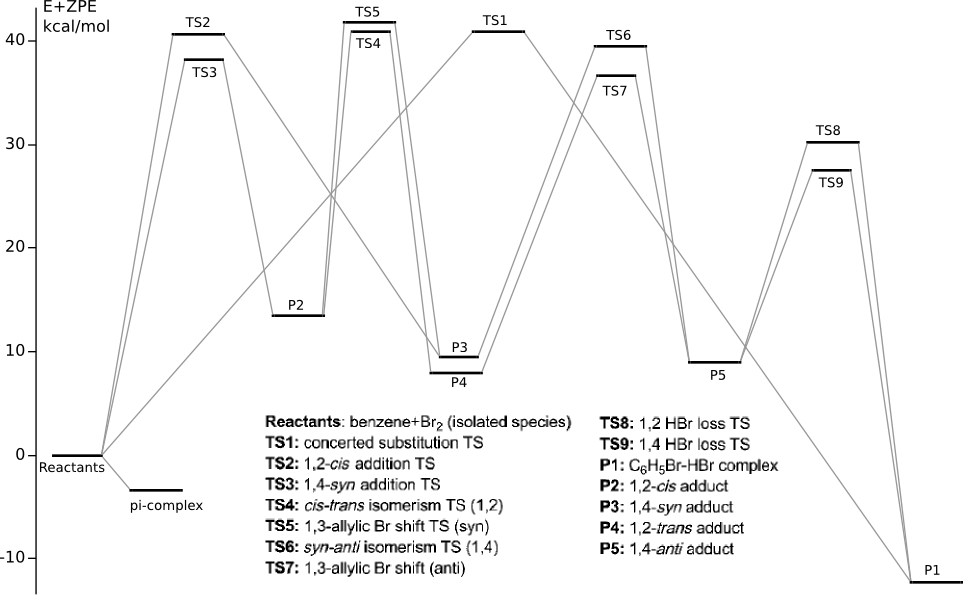
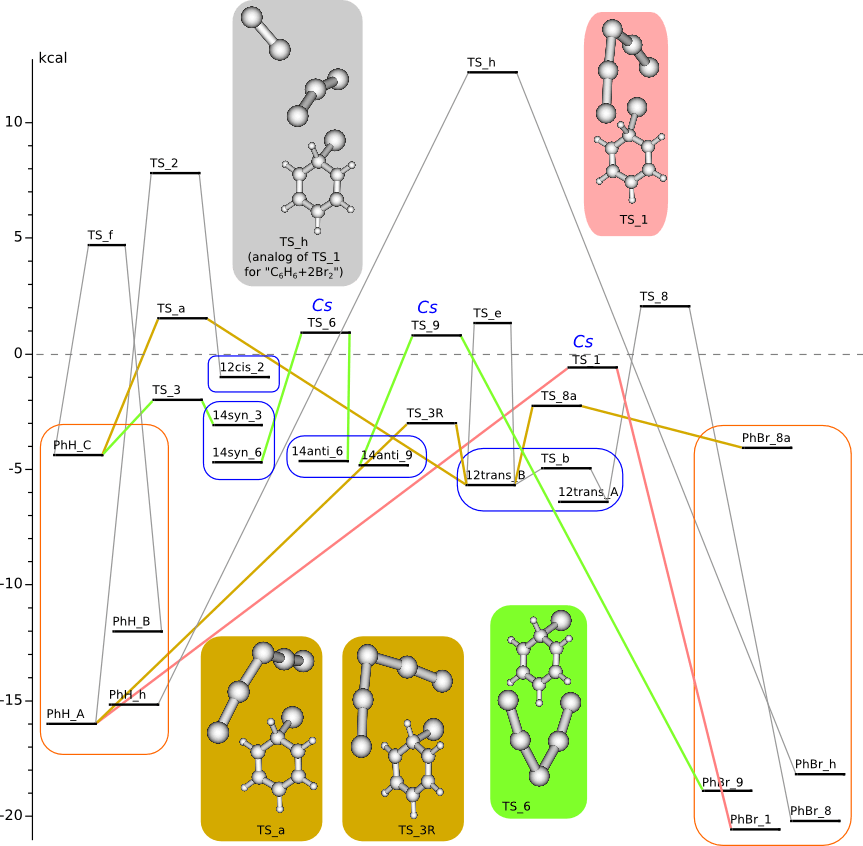
C6H6 + 2Br2 PES
Geometry optimization by DFT/PBE/L1 (PRIRODA program, basis like cc-pVDZ)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
Summary energy of noninteracting reagents is taken to be zero energy.
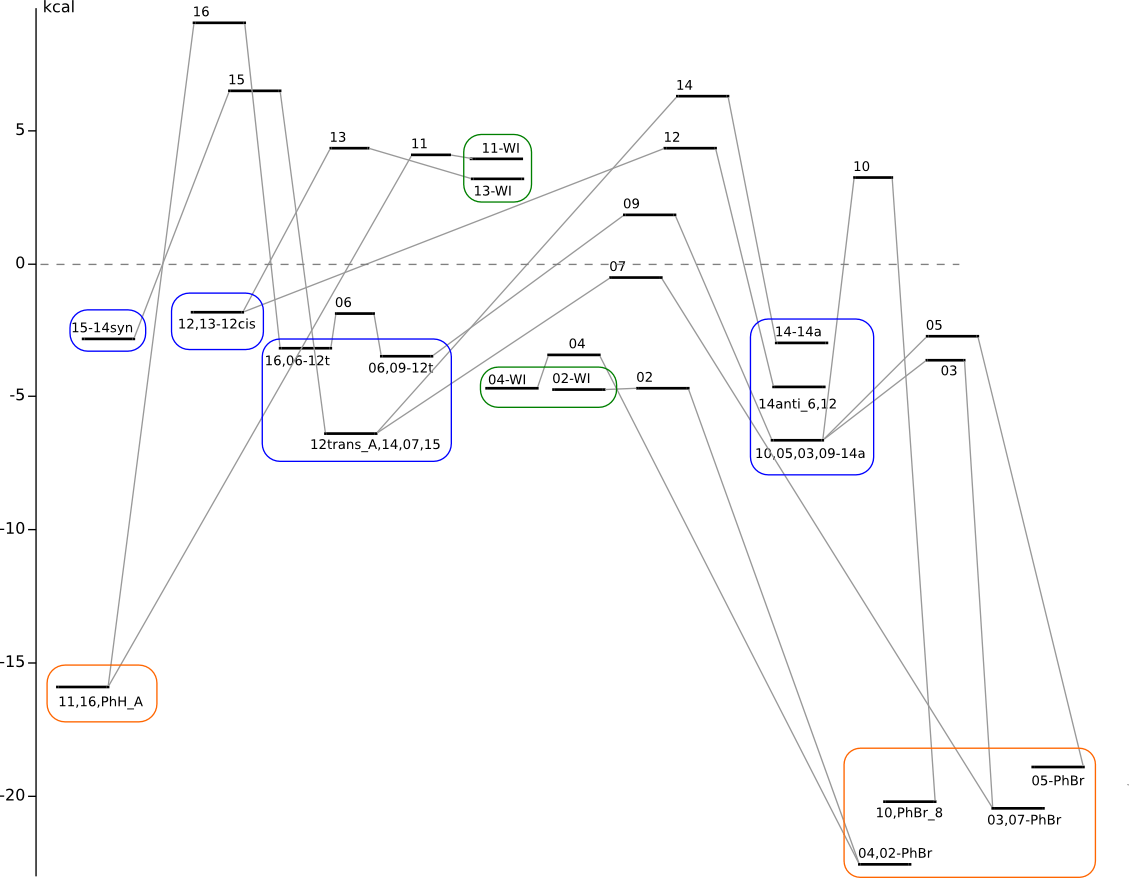
C6H6 + 3Br2 PES
Geometry optimization by DFT/PBE/L1 (PRIRODA program, basis like of cc-pVDZ)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
Summary energy of noninteracting reagents is taken to be zero energy.
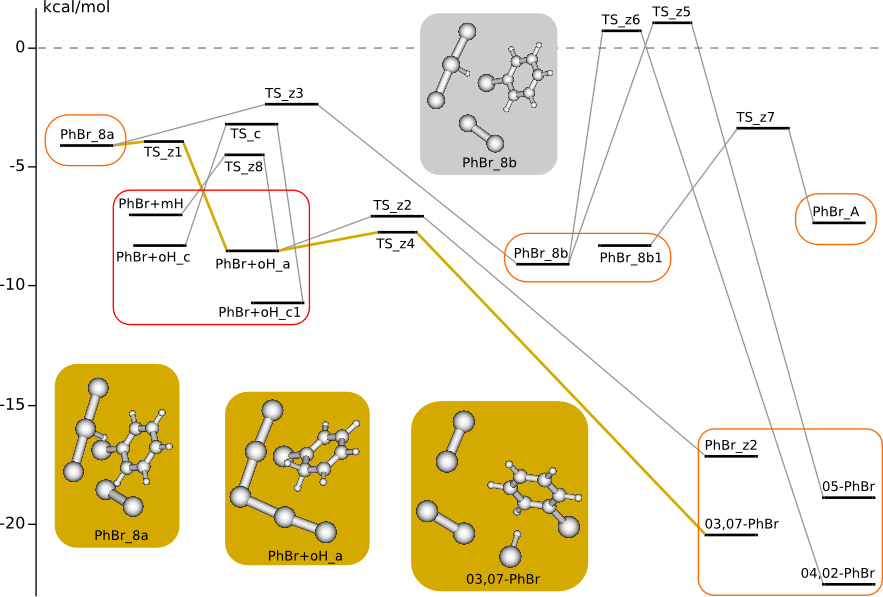
Additional C6H6 + 3Br2 PES
Starting structures for transition states (PhHBr+ + Br5-) was found by Coalescence Kick (CK) method .
CK method: Sergeeva A.P., Averkiev B.B., Zhai H.-J., Boldyrev A.I., and Wang L.-S. J. Chem. Phys. 134, 224304 (2011)
Summary energy of noninteracting reagents is taken to be zero energy.
Relaxation of high-energy complex PhBr_8a (PhBr+HBr3+Br2)
Summary energy of noninteracting reagents is taken to be zero energy.
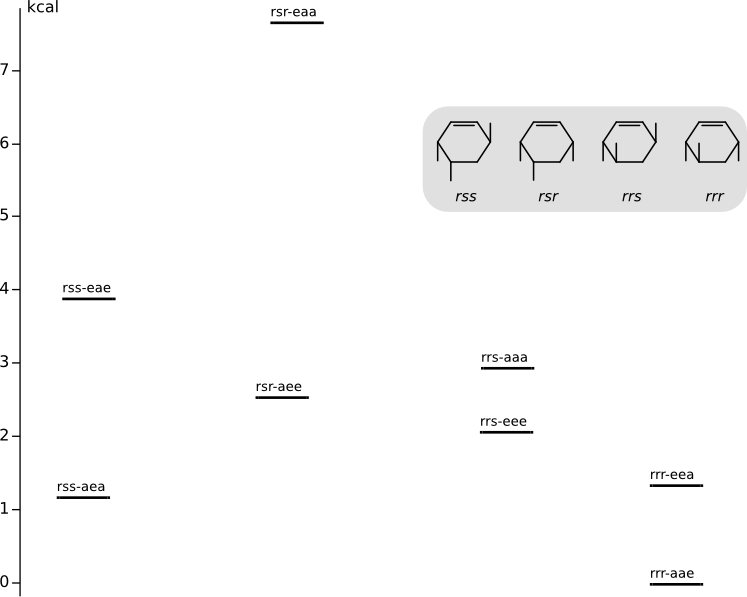
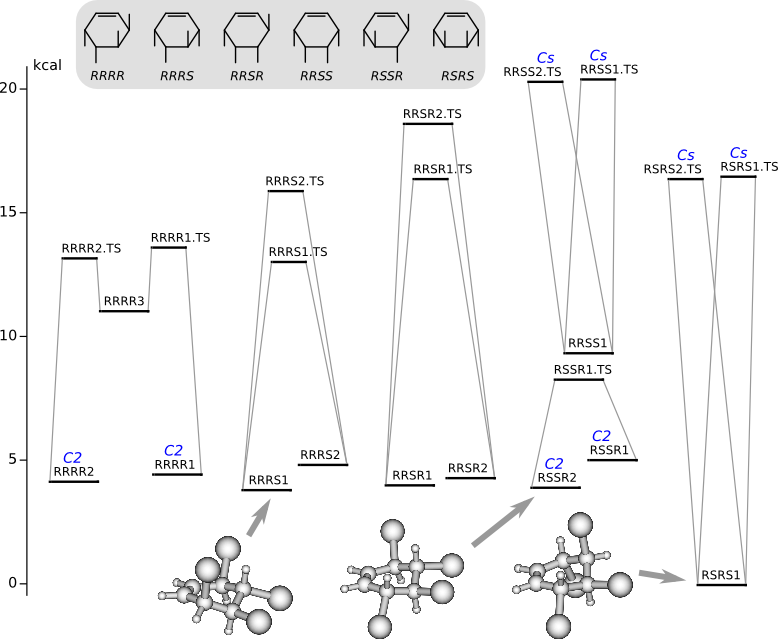
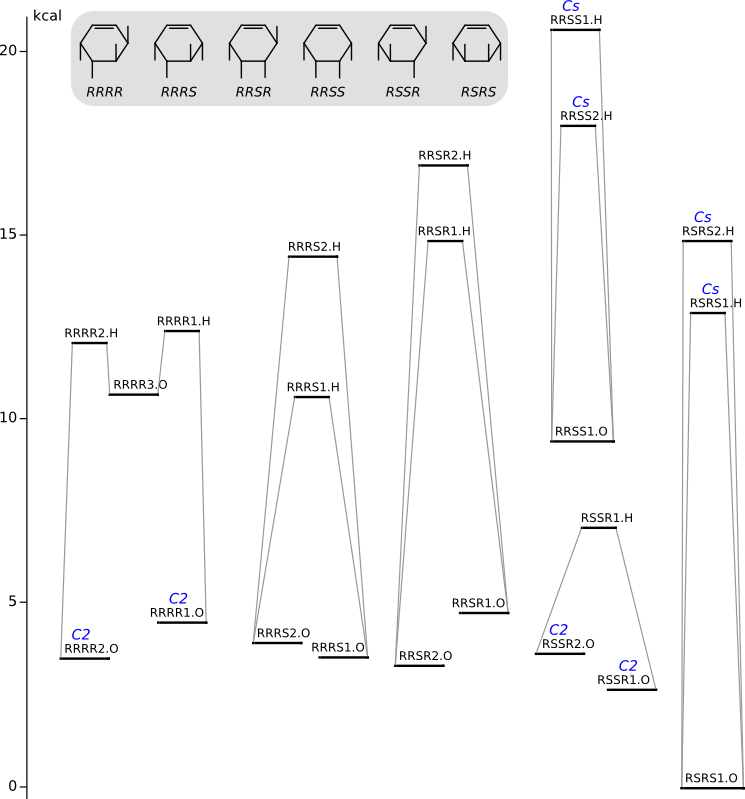
Conformational analysis of 3,4,5,6-tetrabromocyclohexene
Methods of conformational anlysis: MM (Marvin) — MP2 (PRIRODA).
Energy (without ZPE) by riMP2/L1 (cc-pVDZ, PRIRODA program)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
Methods of conformational anlysis: MM (Marvin) — DFT (PRIRODA).
Energy (without ZPE) by DFT/PBE/L1 (cc-pVDZ, PRIRODA program)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
Conformational analysis of 3,4,6-tribromocyclohexene
Energy (without ZPE) by riMP2/L1 (cc-pVDZ, PRIRODA program)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
|