|
|
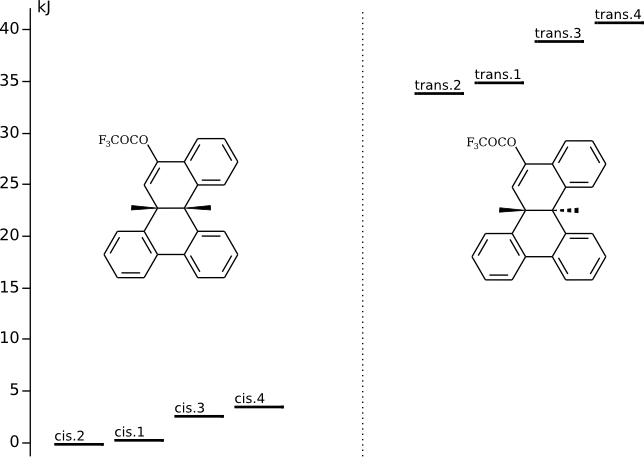
Conformational analysis of 8b,14b-диметил-8b,14b-дигидробензо[g]хризен-10-илтрифторацетат
Methods of conformational anlysis: MM (Marvin+Vconf+Tinker) — DFT (PRIRODA).
Energy (without ZPE) and chemical shifts by DFT/PBE/L22 (cc-pCVTZ) (PRIRODA program)
On figure(s) below click on the level title to download xyz file
with calculated (DFT/PBE/L22) chemical shifts (5-th column) |
==> |
| Click on energy level to view 3D structure (run jmol java-applet) |
==> |
| 
|
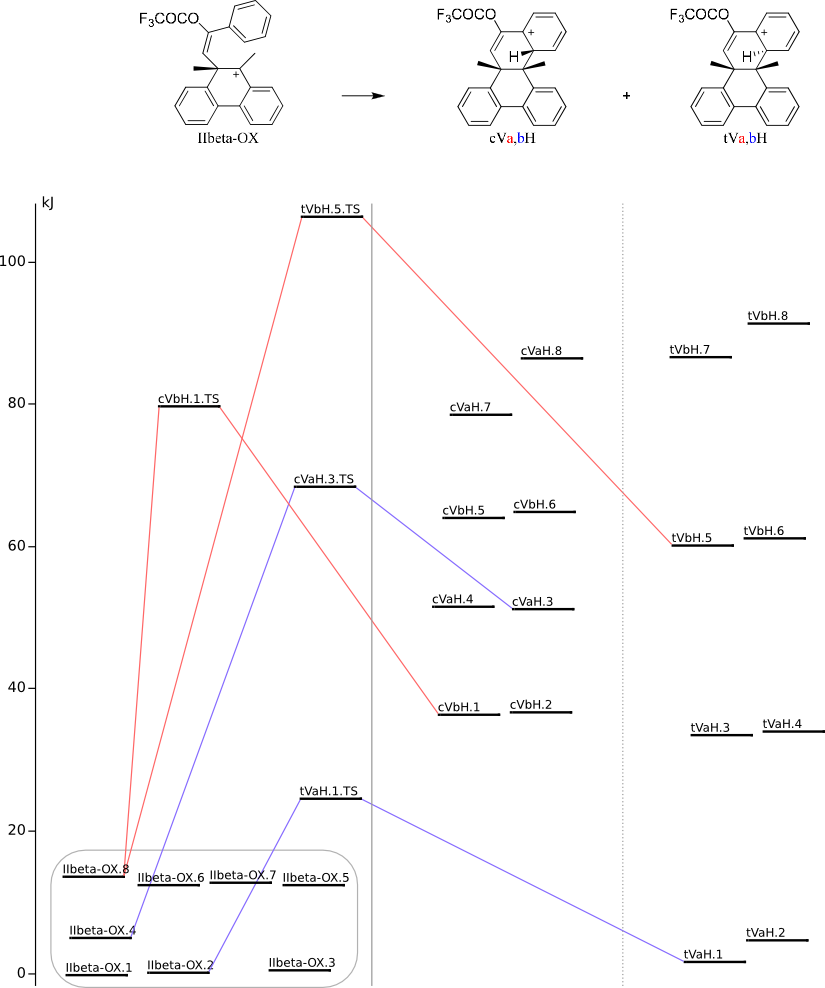
Cyclization into 8b,14b-диметил-8b,14b-дигидробензо[g]хризен-10-илтрифторацетат-H+
Methods of conformational anlysis: MM (Marvin+Vconf+Tinker) — MOPAC (RM1) — DFT (PRIRODA).
Energy (without ZPE) by DFT/PBE/L1 (cc-pVDZ) (PRIRODA program)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
Cyclization into 5a,6-dimethyl-4-phenyl-5,6-dihydro-4H-acephenanthrylene-4,6-bis(ylium) dication
Energy (without ZPE) by DFT/PBE/L1 (cc-pVDZ) (PRIRODA program)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|

Rearrangements of 5a,6-dimethyl-4-phenyl-5,6-dihydro-4H-acephenanthrylene-4,6-bis(ylium) dication
Energy (without ZPE) by DFT/PBE/L1 (cc-pVDZ) (PRIRODA program)
| On figure(s) below click on the level title to download xyz file |
==> |
| Click on energy level to view 3D structure in browser (run JSmol) |
==> |
| 
|
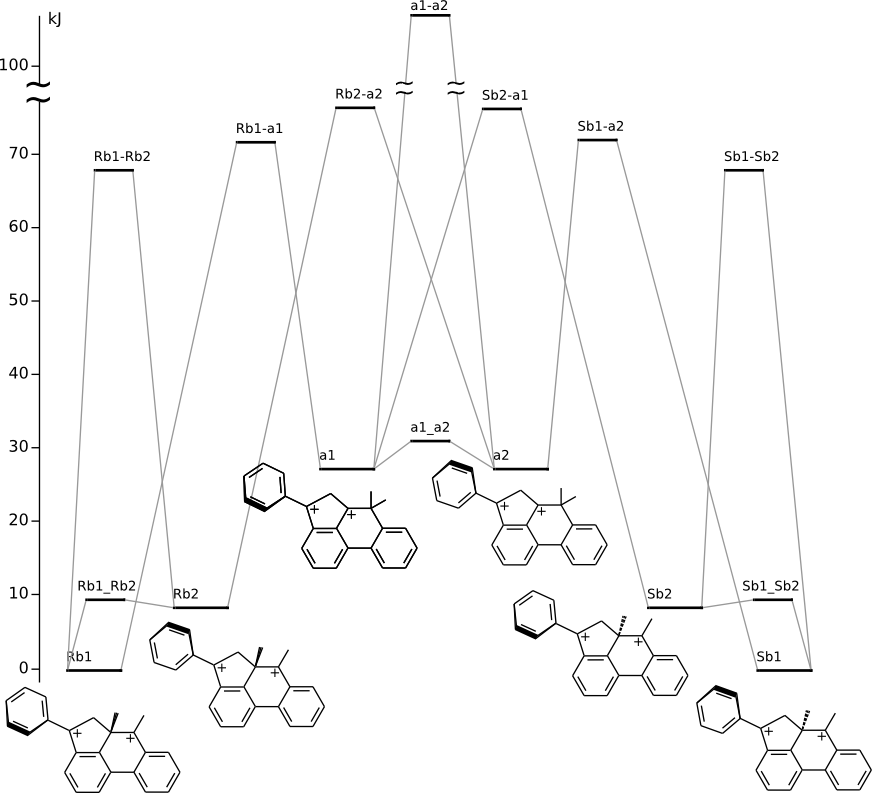
Animation Rb1-a1-a2-Sb1 (Me shift)

Animation Rb1-a1-Sb2-Sb1 (Me shift)

Animation Rb1--Rb2 (Ph rotation)

This study was supported by the Russian Foundation for Basic Research (project no. 13-03-004)
|